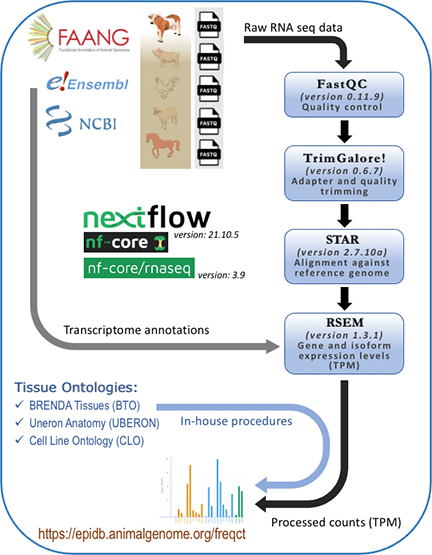
Software and pipeline
Workflow
Data sources
Shell script to run the piepline
#!/usr/bin/bash
#SBATCH -o "stdout.%j"
#SBATCH -e "stderr.%j"
#SBATCH --mem 100GB
##SBATCH -p amd
#SBATCH -N 1
#SBATCH -n 20
#SBATCH -t 12:00:00
GENOME=.../Bos_taurus.ARS-UCD1.2.dna.toplevel.fa.gz
GTF=.../Bos_taurus.ARS-UCD1.2.107.gtf.gz
module load nextflow
export NXF_SINGULARITY_CACHEDIR=/path/NXFContainers/
nextflow run nf-core/rnaseq --input sample_sheet.csv --outdir star_rsem_output --fasta ${GENOME} --gtf ${GTF} -resume -profile singularity --aligner star_rsem
Outcomes
The end results of processed TPM counts can be visulized as
bar graphs showing expression levels (counts) by tissues and genes with our
in-house tool.
* All data use at the EpiDB follow the FAANG data sharing statement.